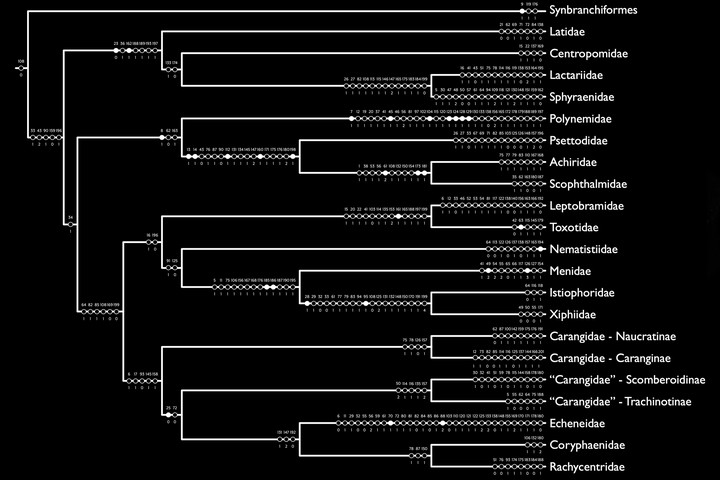
As an intern at The Field Museum, I began to recognize the disagreement between genotypic- and phenotypic-based phylogenies and started integrating datasets of different character types to answer questions about the evolution of fishes. My work initially used Sanger-based approaches to collect molecular data and now employs high-throughput techniques to sequence loci via genome skimming (i.e., mitochondrial genomes) or target enrichment (i.e., ultraconserved elements). I also employ bioinformatic tools to integrate modern DNA datasets with legacy loci to leverage and build upon these abundantly available data. My approaches to capturing morphological data have similarly expanded, beginning with dry skeletal and wet, stained specimens to now include µCT scanning of hard and soft tissues. Today, I integrate and analyze multiple character types to gain a greater understanding of fish evolution and arbitrate phylogenetic discrepancies. I have used my diverse toolkit to generate many evolutionary hypotheses for fishes, including the first phylogeny for the order Carangiformes (billfishes, flatfishes, and jacks), based on genotypic and phenotypic characters. More studies using this integrative approach are in progress, with some representative publications listed above. Check out my publications for more information and representative publications.
Systematics and evolution of fishes